Welcome to the first entry in the Learn by Implementation series. The purpose of this series is to help learn about how to work with the tripod that is, Nd4j, Canova, and Dl4J, by working through an implementation. Generally the code being implemented will be a known, and well understood algorithm (usually Machine Learning related), but I won’t hold myself to such strict rules. Whatever follows this post in the series will always focus on explaining and exploring different aspects of the tripod. The typical flow will include, introducing the algorithm, walking through the main aspects of it, and then programming it. The programming side will go into lengths about various features of the three libraries (whichever are needed for the project), and discuss design decisions.
If you haven’t set up Deeplearning4j, check out this tutorial first!
K-Nearest Neighbor (KNN)
- Overview of KNN
- Designing KNN
- Implementing KNN
- Conclusion
K-Nearest Neighbor, a straight forward classifier, makes for an excellent candidate to start our series on. KNN is known as a “lazy learner” or instance based learner. We are using the term learner pretty loosely here, especially in the wake of DL4J and all of the latent modeling available out of the box.
THE WALK THROUGH
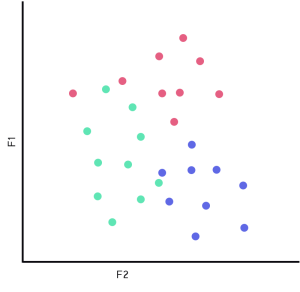
Essentially, KNN’s model is a training set that is used as a look up table to find the k nearest neighbors based on some sort of distance measure. Then, with the k neighbors, apply some sort of classification strategy. In the case below, we will only have two features(dimensions) for simplicity sake, F1 and F2.
FIT THE MODEL
In the case of KNN’s, and all instance based learners, we don’t pre-compute a model/interpretation of the data. Instead, we just store the data to be used on prediction evaluation.
KNN model in 2D (Fig 2.0)
PREDICT
Imagine we have a dataset that corresponds to the image above (Fig 2.0). Where we have 2 dimensions of data, and 3 different labels (represented by red, green, and blue); this data will operate as the KNN’s model. Since we are working with a lazy learner, we can jump right to looking at what happens when we give that classifier and unseen piece of information, known as input.
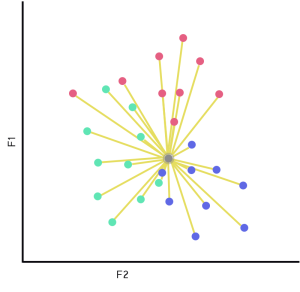
New Input (Fig 2.1)
The grey dot with an aura represents our input. The first step of KNN is to measure the distance from our input against each record we’re holding on to. This acts as the lazy learning part, as the model used to solve the problem is built on request. After we’ve successfully measured the distances to all of the data points, we can select the closest K records.
For this example we are going to use Euclidean for our distance measure; though this could be literally any distance function (that is sensible).
Once we have the neighborhood of k nearest, we need to apply a evaluation strategy. We can take the highest representation of classes (winner takes all), we could add weights based on distance or some other measurement to the class labels, or really just about anything. We’re not going to spend much time deliberating on the concept, but typically the evaluation strategy you’d pick depends on the distribution of the data. All of the approaches will generally have advantages and disadvantages.
With that said, we’re going to apply a winner takes all strategy. It’s easy to understand, and easy to program. If we look at our neighborhood (4 blue, 3 green, 1 red), we can see that we have the largest representation of the blue label (or class). Thus we will have KNN predict blue in this example.
Apply Winner Takes All (Fig 4.0)
There’s not much else to say about KNN, it can be surprising effective for certain tasks/data. There is a lot you can do to increase it’s robustness, and an algorithm like this definitely has it’s place. An algorithm like this will surely fail on outlier examples, or unrepresented cases; with that said, let’s move implement the algorithm and explore Nd4j!
LET’S IMPLEMENT!
public void fit(Dataset dataset); public int[] predict(INDArray input);
public class KNN {
DataSet data;
BaseAccumulation distanceMeasure;
int numberOfNeighbors;
}
Interfaces
- Op offers the ability to take an INDArray x(remember this can be of any size and dimension) and have it operate over another INDArray y, and store said operation in INDArray z, it’s buffer result. An example of an Op would be softMax or finding the index of the max value of an INDArray.
- Accumulation, a sub-interface of Op, look to accumulate results over the operation. So for example the Mean, or even more relevant for us, distance functions such as Euclidean and Manhattan.
Constructors
/**
* Instantiates KNN with the default set up. K=5, distanceMeasure = Euclidean
*/
public KNN() {
this(5);
}
/**
* Instantiates KNN with Euclidean as it's distanceMeasure and a user set neighborhood size.
* @param numberOfNeighbors - The number of neighbors to look at, also known as K.
*/
public KNN(int numberOfNeighbors) {
this(numberOfNeighbors, new EuclideanDistance());
}
/**
* Instantiates KNN with the specified distanceMeasure and neighborhood size.
* @param numberOfNeighbors - The number of neighbors to look at, also known as K.
* @param distanceMeasure - The kind of distance measurement. (Euclidean, Manhatten, ...)
*/
public KNN(int numberOfNeighbors, BaseAccumulation distanceMeasure) {
this.distanceMeasure = distanceMeasure;
this.numberOfNeighbors = numberOfNeighbors;
}
There isn’t too much to say about these constructors. We’re just given the user the opportunity to configure his KNN to his needs. Moving on…
Internal Methods – support for the public
We’ll need a way to measure the distance from our input to each of the records in our dataset (model). We’ll call the method measureDistance and define it as the following:
/** * Takes a feature and compares the distance between itself and each feature in a matrix. * * @param feature - The feature to measure against the matrix/ * @param featureMatrix - The matrix of data, likely from a dataset. * @return a vector of distances that line up with the rows of the featureMatrix. */ private INDArray measureDistance(INDArray feature, INDArray featureMatrix, BaseAccumulation distanceMeasure);
We can see that we pass in a feature (or multiple features), feature being our input. We pass in our featureMatrix, which is the data from the dataset that we have stored in memory. Lastly, we specify the distance measurment. This defines how we want to measure between each set of records and the input. We expect to return a vector for each input passed in. So if our INDArray for the feature has more than one record, we will have more than one vector returned (as a matrix).
Once we have the distance measurements for each record in our model (the dataset we have in memory), we need to find the neighborhood that we are interested in. We’ll call this method findKNeighbors and define at as the following:
/** * Pairs up the measured distances for each observation with the labels. * Sorts them on the distance metric, and grabs the k smallest distance records. * * @param k - the size of the neighborhood. * @param returnDistanceMeasure - Whether we should include the measure in our * neighborhood information schema. * @param distanceNDArray - The measured distance vector. Needs to be the * same length as the number of rows for labelMatrix. * @param labelMatrix - The label matrix of the dataset at hand. * @return Returns the neighborhood matrix, comprised of distance and labels. * dims are [0 = the input example, 1=instances in the neighborhood * running 0 to k, 2= distance and labels * (index 0 is distance measure, the remaining are labels) ] */ private INDArray findKNeighbors(int k, boolean returnDistanceMeasure, INDArray distanceNDArray, INDArray labelMatrix);
The result of this method should give us a INDArray of the k nearest neighbors for each set of inputs to exist. The structure will look like something below. Typically, dim0 refers to rows, dim1 refers to columns, and dim2 would be our 3rd dimension(Credit: Alex Black). I’ve just orientated the view below differently.
findKNeighbors return structure (Fig 5.0)
For the parameters, k refers to the size of the neighborhood. The boolean, returnDistanceMeasure, determines if we should expect to have the measurements included with the returned neighborhood.
distanceNDArray will be the output of the measureDistance method. It will either be a vector or a matrix (if more than one input was passed through to the predict method). Lastly, there is the labelMatrix, which should be a matrix of labels that is served up from the same place that served up the featureMatrix in the measureDistance method.
Implement Fit(…)
This boils down to just being a setter for the KNN. We will just store the dataset in memory for the given instance of KNN.
/**
* KNN doesn't learn anything intrinsic about it's data. Instead, on predict call; each record in the set will be looked at.
* This model employs Lazy Learning/Instance-based Learning.
* @param data - Expected to be as much relevant data as possible.
*/
public void fit(DataSet data) {
this.data = data;
}
Implement Predict(…)
The first thing we need to do for our predict method, is to get some variables initialized.
/**
* Takes in n rows of input records and spits out n predictions.
* NOTE: There may be room for optimization in handling the multi-record input.
* @param input - The matrix (or single vector) of input to test.
* @return a corresponding array of label index predictions.
*/
public int[] predict(INDArray input) {
INDArray features = data.getFeatures();
INDArray labels = data.getLabels();
int numberOfInputs = input.rows();
int[] predictions = new int[numberOfInputs];
boolean includeMeasure = false;
int offset = (includeMeasure) ? 1 : 0;//Hardcoded offset, this represents that column is used for distance in the findKNeighbors method.
...
We’ll grab the feature matrix from the model Dataset we have in memory. In a separate INDArray labels, we will grab the matrix of label values from the model Dataset. We want to capture the number of rows in our input. Lastly we want to allocate an array of ints, predictions, to the size of the number of rows in our input.
Now that we’ve allocated and set up the variables we will need, we can make our first call to measureDistance(…)
INDArray distanceNDArray = measureDistance(input, features, distanceMeasure);
Before we jump into this method, we can take a moment to look at what we are passing in. All of the values are from above, except distanceMeasure. distanceMeasure is a value we declared up in our class variable section. It represents a distance measurement for our purposes. The default construction of this class will utilize EuclideanDistance.
Detour -> Implement measureDistance(…)
We’re taking a quick detour into implementing the measureDistance(…) method, afterwards we’ll hop back into implementing predict(…). The first thing we’ll do, like always, is initialize the variables we need for the remainder of the method.
/**
* Takes a feature and compares the distance between itself and each feature in a matrix.
*
* @param feature - The feature to measure against the matrix.
* @param featureMatrix - The matrix of data, likely from a dataset.
* @return a vector of distances that line up with the rows of the featureMatrix.
*/
private INDArray measureDistance(INDArray feature, INDArray featureMatrix,
BaseAccumulation distanceMeasure) {
int numberOfRows = featureMatrix.rows();
int numberOfInput = feature.rows();
INDArray distances = Nd4j.zeros(numberOfRows, numberOfInput);
We’ll initialize a variable numberOfRows, and store the row count from the featureMatrix parameter. We also will acquire the numberOfInput from the feature parameter. Lastly, we’ll instantiate a new empty INDArray of the dimensions numberofRows, numberofInput (maps to a matrix: rows, columns) called distances. We’ll store our measurements in here.
ND4J – A Factory
When we look to create a new INDArray, we access the class Nd4j.staticMethod(…) to instantiate a new N-dimensional array. From here we have a few options to carry our task out. Below is just a light overview of the create(…) method, but be sure to take a look for yourself in your favorite IDE and experiment to get a handle on the process.
- Nd4j.create(…) – There are quite a few implementations of this method, some where you pass in an array of values (double or float). Whenever you do pass in an array of data, you are also given the choice of shaping each of the dimensions. There are also options to create an Nd4j object by specifying the rows and columns as ints, or more dimensions as needed.
Now that all of our information needed is initialized, we can move onto the operational part of the method.
for (int inputIndex=0; inputIndex < numberOfInput; inputIndex++) {
distanceMeasure.setX(feature.getRow(inputIndex));
for (int rowIndex=0; rowIndex < numberOfRows; rowIndex++) {
distanceMeasure.setCurrentResult(0);
distanceMeasure.setY(featureMatrix.getRow(rowIndex));
Nd4j.getExecutioner().execAndReturn(distanceMeasure);
distances.put(rowIndex, inputIndex, distanceMeasure.currentResult());
}
}
return distances;
}
We set up a double for loop. The first loop will iterate over each input feature that we have. You can see that we are setting the X value for distanceMeasure to the current input feature. The second for loop iterates over the featureMatrix (KNN model) and measures the distance between the current row in the featureMatrix and the current input feature. For each iteration in this loop, we clear our accumulation value (we wouldn’t want to use a previous calculation and have it count towards our current calculation), set the Y value for the distanceMeasure, and then tell the executor to execute and return the value. Our last step is to store the result of the distance calculation into our distances INDArray.
Once we’ve finished our double loop, we simply return our distances INDArray.
End Detour <- Back to implementing Predict
/**
* Takes in n rows of input records and spits out n predictions.
* NOTE: There may be room for optimization in handling the multi-record input.
* @param input - The matrix (or single vector) of input to test.
* @return a corresponding array of label index predictions.
*/
public int[] predict(INDArray input) {
INDArray features = data.getFeatures();
INDArray labels = data.getLabels();
int numberOfInputs = input.rows();
int[] predictions = new int[numberOfInputs];
boolean includeMeasure = false;
int offset = (includeMeasure) ? 1 : 0;//Hardcoded offset, this represents that column is used for distance in the findKNeighbors method.
INDArray distanceNDArray = measureDistance(input, features, distanceMeasure);
Alright back on track with the predict method. If you remember (or you can expand the above code) that we just called measureDistance(…) and stored the result in an INDArray distanceNDArray. Our next step is going to be to be making a call to our other private method, findKNeighbors(…).
//This is a 3d array INDArray nearestNeighbors = findKNeighbors(numberOfNeighbors, includeMeasure, distanceNDArray, labels);
We pass in our neighborhood size, a boolean that specifies if we want the distance measure included with the neighborhood results, the distanceNDArray (which was returned from the measureDistance(…) call), and the labels (a matrix of labels).
Detour -> Implement findKNeighbors(…)
As we discussed earlier, findKNeighbors(…) goal is to capture the K closest neighbors based on some measurement. We’ll start by initializing the necessary variables and then move onto the operational part of the code.
/**
* Pairs up the measured distances for each observation with the labels. Sorts them on the distance metric, and grabs the k smallest distance records.
*
* @param k - the size of the neighborhood.
* @param returnDistanceMeasure - Whether we should include the measure in our neighborhood information schema.
* @param distanceNDArray - The measured distance vector. Needs to be the same length as the number of rows for labelMatrix.
* @param labelMatrix - The label matrix of the dataset at hand.
* @return Returns the neighborhood matrix, comprised of distance and labels.
* dims are [0 = the input example, 1=instances in the neighborhood running 0 to k, 2= distance and labels (index 0 is distance measure, the remaining are labels) ]
*/
private INDArray findKNeighbors(int k, boolean returnDistanceMeasure, INDArray distanceNDArray, INDArray labelMatrix) {
//Horrizontally merge the vector with the Matrix.
int labelLength = labelMatrix.columns();
int distLength = (returnDistanceMeasure) ? 1 : 0;
int infoSize = distLength + labelLength;
INDArray mergedSet;
INDArray kNeighbors3d = Nd4j.create(distanceNDArray.columns(), k, infoSize);
//Create the indexes that decide what info we are including the distance measure or not
int[] indexes = new int[infoSize];
for (int i=0; i < indexes.length; i++)
indexes[i] = i + (1 - distLength);
...
We store the number of labels as labelLength, we check if the returnDistanceMeasure is true or false and assign a 1 or 0. We then create a variable infoSize, which will tell us the number of columns we expect to return per input feature. We instantiate mergedSet; which will be used in the next part. Additionally, we allocate space for a 3d INDArray, kNeighbors3d. Lastly, we create an array of ints that will determine our column view port for the returning structure.
Now onto the operational code…
for (int z = 0; z < distanceNDArray.columns(); z++) {
mergedSet = Nd4j.hstack(distanceNDArray.getColumn(z), labelMatrix);
INDArray sortedSet = Nd4j.sortRows(mergedSet, 0, true).getColumns(indexes);
INDArray kNeighbors = kNeighbors3d.tensorAlongDimension(z, 2, 1);
for (int i=0; i < k; i++) {
kNeighbors.putRow(i, sortedSet.getRow(i));
}
}
return kNeighbors3d;
}
Here we do a double for loop. The first loop iterates over the number of inputs, or z dimension. Then we merge the distance vector for the given input (z) with the labelMatrix, into one matrix. We take our mergedSet and sort the rows ascending based on the distance metric using a sortRows method(more on that later). After we’ve sorted, we make a call to getColumns(…), this allows us to create a view that will include or exclude the distance metric from the return result.
We also use a function on the INDArray class called tensorAlongDimension(…). In short, this allows us to take an N-dimensional array and create a new view of it. In this case we are taking a 3d Array and projecting it to a 2d matrix over at a specific position on the z dimension (0 for our case). What’s neat is that not only does this function increase our flexibility in N-dimensions, but when you make a change to this “view”, it affects the underlying data structure. Now that we have a 2d matrix for a specific slice in our 3d array, we can iterate over our sorted matrix view and copy over the closest k values.
End Detour <- Back to implementing Predict
Now that we’re finished setting up our two private methods, we can finish up our predict(…) method.
//Collect up the label columns, offset represents our distanceVector column
//dimension 2 refers to distanceMeasure & labels, also referred to as info.
int[] indexes = new int[nearestNeighbors.size(2) - offset];
for (int i=0; i < indexes.length; i++) {
indexes[i] = i + offset;
}
Basically what we are doing is collecting up the column indexes of the labels and excluding the measure data if present. So if we had 3 labels and a measure distance, our measure distance would reside in index 0, and indexes 2, 3, and 4 would contain labels. If we set includeMeasure to false at the start of predict(…), we don’t have to worry about filtering the measure distance out
Now that we’ve collected up the label indexes, we can do the following
for (int i=0; i < numberOfInputs; i++) {
//Increment our index that refers to the input index
//Grab the largest count index of the label columns in of the k
//nearestNeighors for the ith input
INDArray measureAndLabelMatrix = nearestNeighbors.tensorAlongDimension(i, 2, 1);
INDArray labelMatrix = measureAndLabelMatrix.getColumns(indexes);
int predictedLabelIndex = Nd4j.getBlasWrapper().iamax(Nd4j.sum(labelMatrix, 0));
predictions[i] = predictedLabelIndex;
}
return predictions;
}
Okay, so lets break this down. We are looping over the number of input features, the things we’re are trying to classify. For each input feature, we are creating a 2d matrix “view” with the method tensorAlongDimension(…) for the specific feature input (remember, the nearestNeighbors structure looked like this; we are iterating over dimension 0).
So the method works like this, tensorAlongDimension(index, dimensions, to, view), where the index refers to the unmentioned dimension. The order of the dimensions to follow is important (refer to this).
Okay so now we have our 2d neighborhood matrix for a given input feature. We then store the data we are interested into another INDArray, labelMatrix. Once we have a view port of our data that contains just labels for the k-neighborhood, we can sum across the rows (dimension 0), and make a call to our BLAS implementation for the index of the largest value in our summed vector. Lastly, we’ll store the result into our predictions array for the current input feature.
Source Code
Closing Comments
This concludes the first in the series of Learn By Implementation. I hope this format was useful, I’ll continue to work on refining the format of this series. With that said, if you have any constructive criticism, questions or what have you, please let me know either in the comments below or at my twitter @depiesml.





[…] See more about this article by clicking the link here: https://depiesml.wordpress.com/2015/09/03/learn-by-implementation-k-nearest-neighbor/ […]
LikeLike
Is there something wrong with the last part of findKNeighbors? You assign values to kNeighbors, but kNeighbors3d always stays zero. I am also not quite sure what tensoring along the empty kNeighbors3d is for – just initializing kNeighbors with the correct dimensions?
LikeLike
The tensorAlongDimension(…) method creates a view. So the resulting structure modifies the original INDArray(kNeighbors is a 2d matrix view of a position in the 3dkNeighbors3d, basically a pointer). I attempted to clarify that in the description of the code. So because I’m expecting potentially more than one input, I have a 3d structure as opposed to just one 2d matrix. Let me know if I need to clarify further.
LikeLike
Ah, I see – thanks.
Found my error: Somehow eclipse autocompleted Nd4j.sort instead of Nd4j.sortRows. Probably because I worked with the same pom.xml as in your “Getting started tutorial”, which uses 0.4-rc0 instead of 0.4-rc1.2.
LikeLike
[…] when needed, and a single buffer INDArray: x, y and z, respectively. In a previous tutorial, LBI: KNN, we used the Euclidean Op, which is a great example of a two-input Op. Lets take a look at how we […]
LikeLike
How do i execute this code. My eclipse is not able to import the header files.
LikeLike
For future readers. The distanceMeasure.setCurrentResult(0); has been deprecated. I think you can just use distanceMeasure.setFinalResult(0); according to the javadoc. Eclipse even suggested the change but just thought I would note the change here.
Thanks for the great tutorial. Always good to start with a kNN example 🙂
LikeLiked by 1 person
Hi
In BaseAccumulation Class there was no method “”setCurrentResult() “” , So how can i set current Result = 0
Iam fresher to deep learning, Could any one help me how to start programming basics ?
Thanks
Narendra k
LikeLike
Hi depiesms22,
It is such a helpful post. I want to use KNN with regression to predict a real value based on its neighbours, can you please guide me for the same?
Thanks in advance.
LikeLike
Hey Psasi, I’m in the middle of putting together a whole new set of tutorials on Machine Learning using Nd4j. However, work is pretty busy right now (as the year ends), and probably won’t start pushing them out until Mid-January. In the meantime, maybe this source will help you in the right direction.
http://www.saedsayad.com/k_nearest_neighbors_reg.htm
Ultimately, I suspect the solution is very dependent on the data you have.
Mike
LikeLike
Thanks a lot Mike.
LikeLike